This Groundbreaking DNA Tool Could Edit-Out Genetic Disease
The DNA-editing tool revealed by researchers today could potentially "fix" the vast majority of known genetic diseases. According to the research text published today, the system of "Prime editing" could potentially "correct about 89% of known pathogenic human genetic variants." That means that most targetable genetic disease-causing bacterium, viruses, and other microorganisms could potentially just be switched out for non-harmful alternatives in the human body.
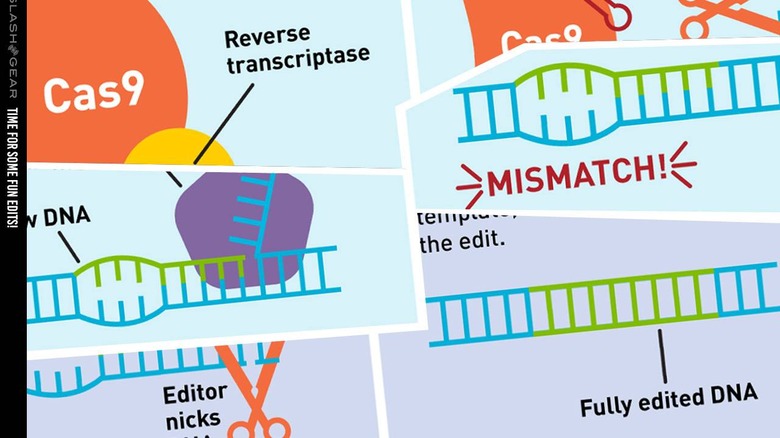
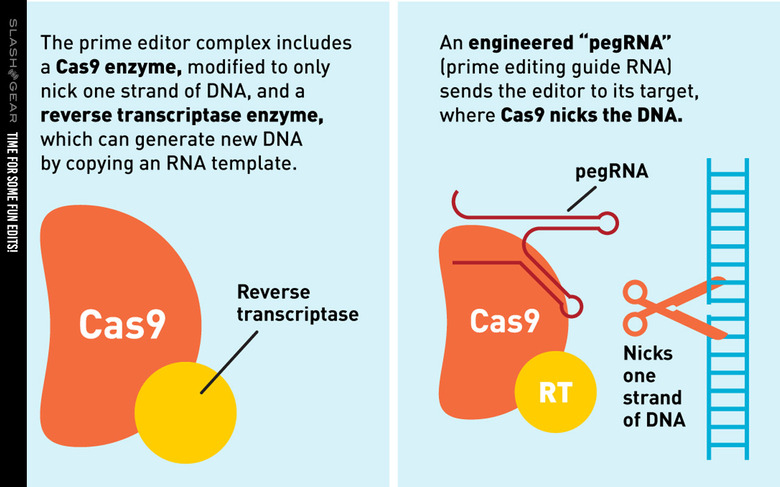
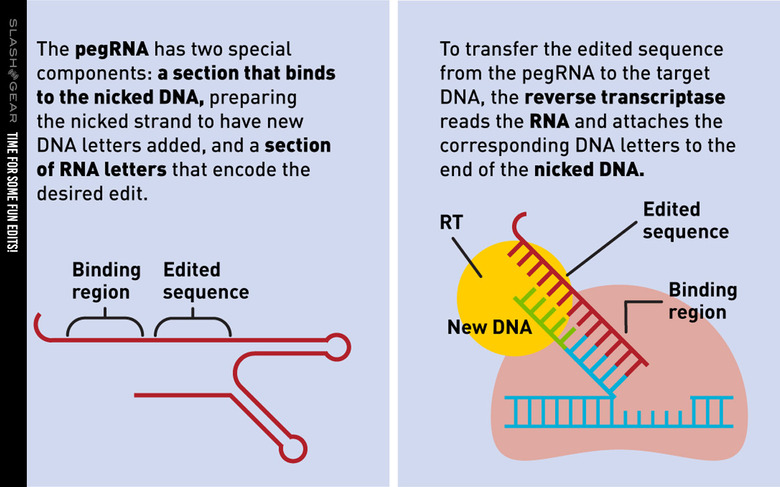
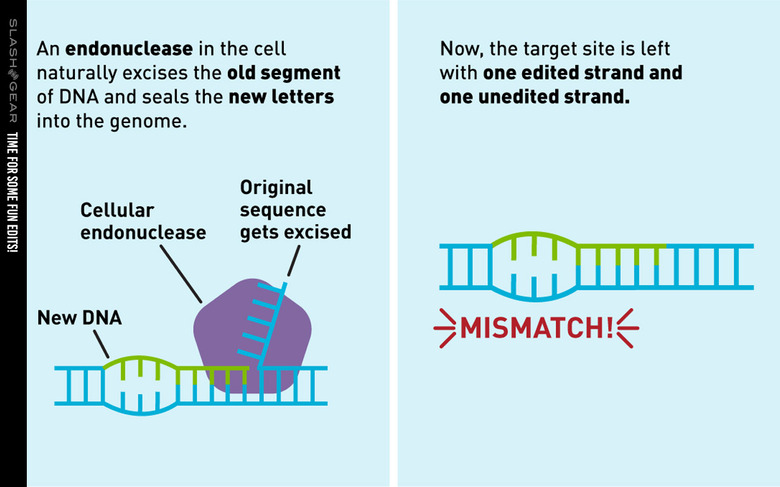
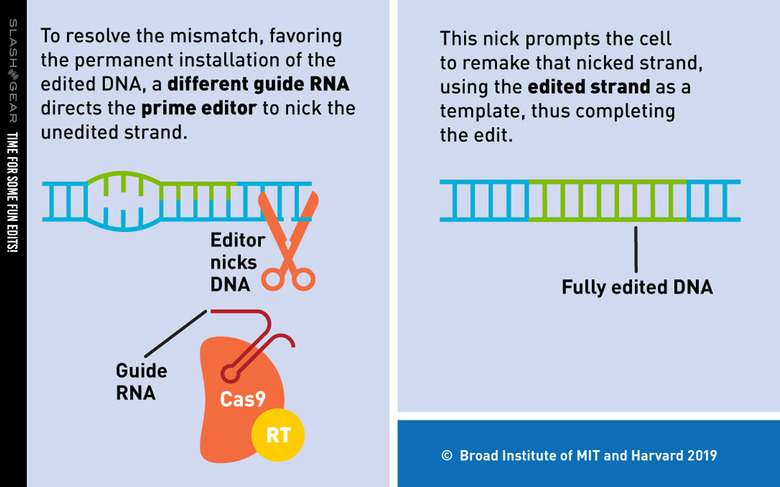
The study of which we speak today was conducted by a team from the Broad Institute of MIT and Harvard. They've created a new genome editing method they say is both versatile and extremely precise. They've created a single machine which uses a CRISPR-Cas9 fused to an engineered reverse transcriptase, programmed with a prime editing guide RNA (pegRNA), to both specify a target site and encode the edit that's needed.
According to the research paper published this week from these scientists and researchers, they've had quite a bit of success in early testing. As they've written, they've "performed more than 175 edits in human cells including targeted insertions, deletions, and all 12 types of point mutation without requiring double-strand breaks or donor DNA templates."
They've shown 4 human cell lines and primary post-mitotic mouse cortical neurons "support prime editing with varying efficiencies." "We're not aware of another editing technology in mammalian cells that offers this level of versatility and precision with so few byproducts," said a statement on the research from several associated leaders, like researcher David Liu.
"The versatility of prime editing quickly became apparent as we developed this technology," said first author on the research, Andrew Anzalone. "The fact that we could directly copy new genetic information into a target site was a revelation. We were really excited."
For more information on this subject, take a peek at the research paper "Search-and-replace genome editing without double-strand breaks or donor DNA." This paper was published to Nature on the 21st of October, 2019. This paper was authored by Andrew V. Anzalone, Peyton B. Randolph, Jessie R. Davis, Alexander A. Sousa, Luke W. Koblan, Jonathan M. Levy, Peter J. Chen, Christopher Wilson, Gregory A. Newby, Aditya Raguram, and David R. Liu, and can be found with code DOI:10.1038/s41586-019-1711-4 as of this afternoon.