11 chameleon species newly split from one
Fooled in the past by color variations, scientists discover the truth about this one panther chameleon: it's actually 11 different species. Using high-resolution color photographs and blood samples from a cool 324 Furcifer pardalis (panther chameleon) individuals along Madagascar's Northern coast, a team of scientists lead by Djordje Grbic from the Laboratory of Artificial and Natural Evolution in the Department of Genetics and Evolution, University of Geneva, Geneva, Switzerland, found that variations were such that one species must split into 11. Upon questioning, these chameleon brethren had no comment on their likely shared ancestry.
While they may well have had a single set of grandparents in the past, today there are 11 different species of Furcifer pardalis, AKA panther chameleon, on the Northern coast of Madagascar. The panther chameleon's entire habitat was explored and samples from the top to the very bottom of its Madagascar-based environment was sampled.
Both mitochondrial and nuclear DNA sequence analyses showed "strong genetic structure among geographically restricted haplogroups." Limited gene flow was discovered here.
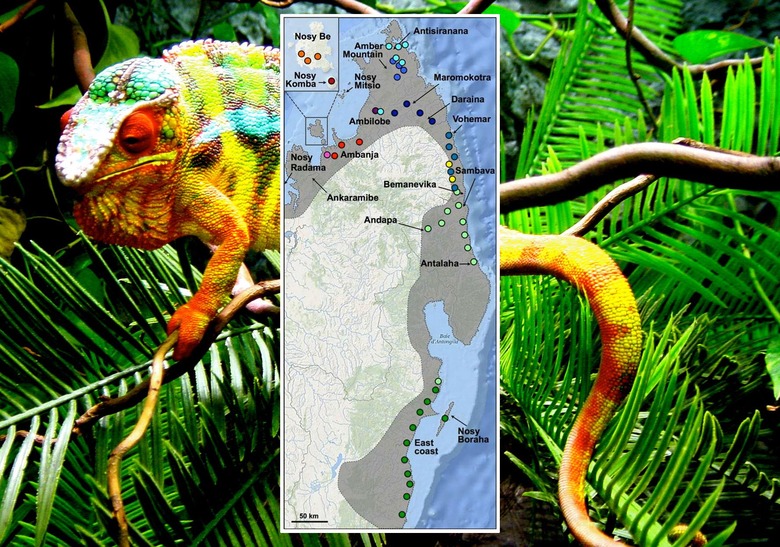
ABOVE: Fig. 1 Sampling panther chameleons in Madagascar. Samplinglocations are indicated with colours corresponding to mito-chondrial haplogroups shown in Fig. 2. The location of a singleindividual from haplogroup 10 (light green) found on the eastcoast probably represents the southern limits of this haplo-group distribution. The most western locations of Nosy Rad-ama and Ankaramibe were not sampled; instead, analysis wasperformed on captive individuals (haplotype L1 in Fig. 2). Theshaded area represents the species entire distribution range.
This extended family of panther chameleon is quickly changing, from generation to generation, changing in such a diverse manner that entirely new species are popping up.
Using five anatomical components as a guide, patterns in 3D color space on these chameleons were used as additional basis for analysis. Using these color pattern guides, this group of scientists "efficiently predict assignment of male individuals to mitochondrial haplogroups."
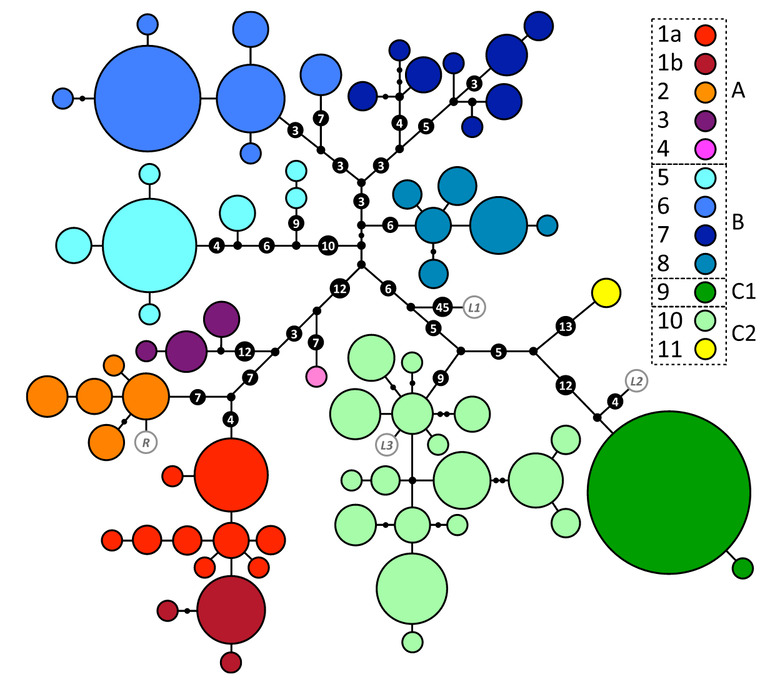
ABOVE: Fig. 2 Panther chameleon mtDNA genea-logical UMP (Cassens et al. 2005) net-work. Haplotypes are represented withcoloured circles, the sizes of which areproportional to the number of individu-als (smallest = 1, largest = 72). Inferredmissing haplotypes are shown as smallblack dots, and each line indicates amutational change (numbers of muta-tions >1 are shown inside larger blackdots). Haplotypes are grouped into col-our-coded haplogroups; sub-haplogroups1a and 1b correspond to samples fromthe northwest coast and the island NosyKomba, respectively. Haplotypes inwhite with grey border circles corre-spond to animals from Reunion (R) andthe pet market (L1–L3).
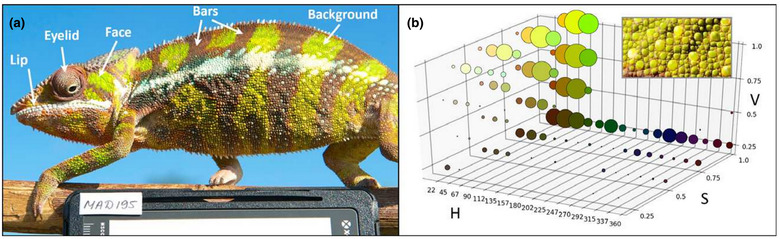
Fig. 3 Colour variation in male panther chameleons.
You can find the full paper in Molecular Ecology under title "Phylogeography and support vector machineclassification of colour variation in panther chameleons" and code doi: 10.1111/mec.13241. This paper was authored by Djordje Grbic, Suzanne V Saenko, Toky M Randriamoria, Adrien Derby, Achille P Raselimana, and Michel C Milinkovitch.
BONUS: The team converted their results into a "simple visual classification key" that they're making available to local trade managers to "avoid local population overharvesting."
This "harvesting" they speak of is referring mostly to capture and transport for pets for humans, not so much for their lesser-known magical abilities.
